Variants in CTSB likely drive the association with PD and are associated with CTSB expression in multiple brain regions
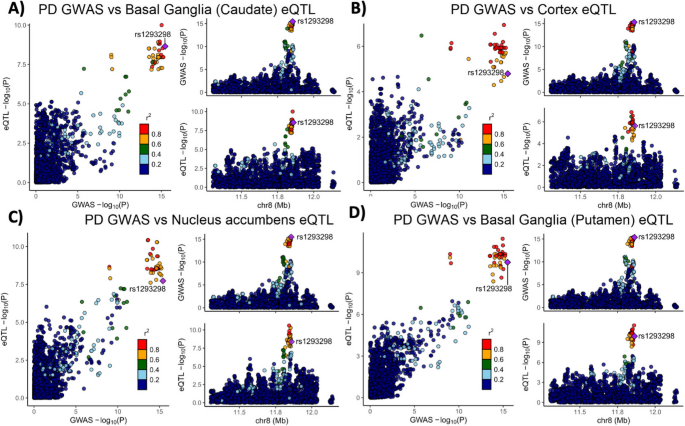
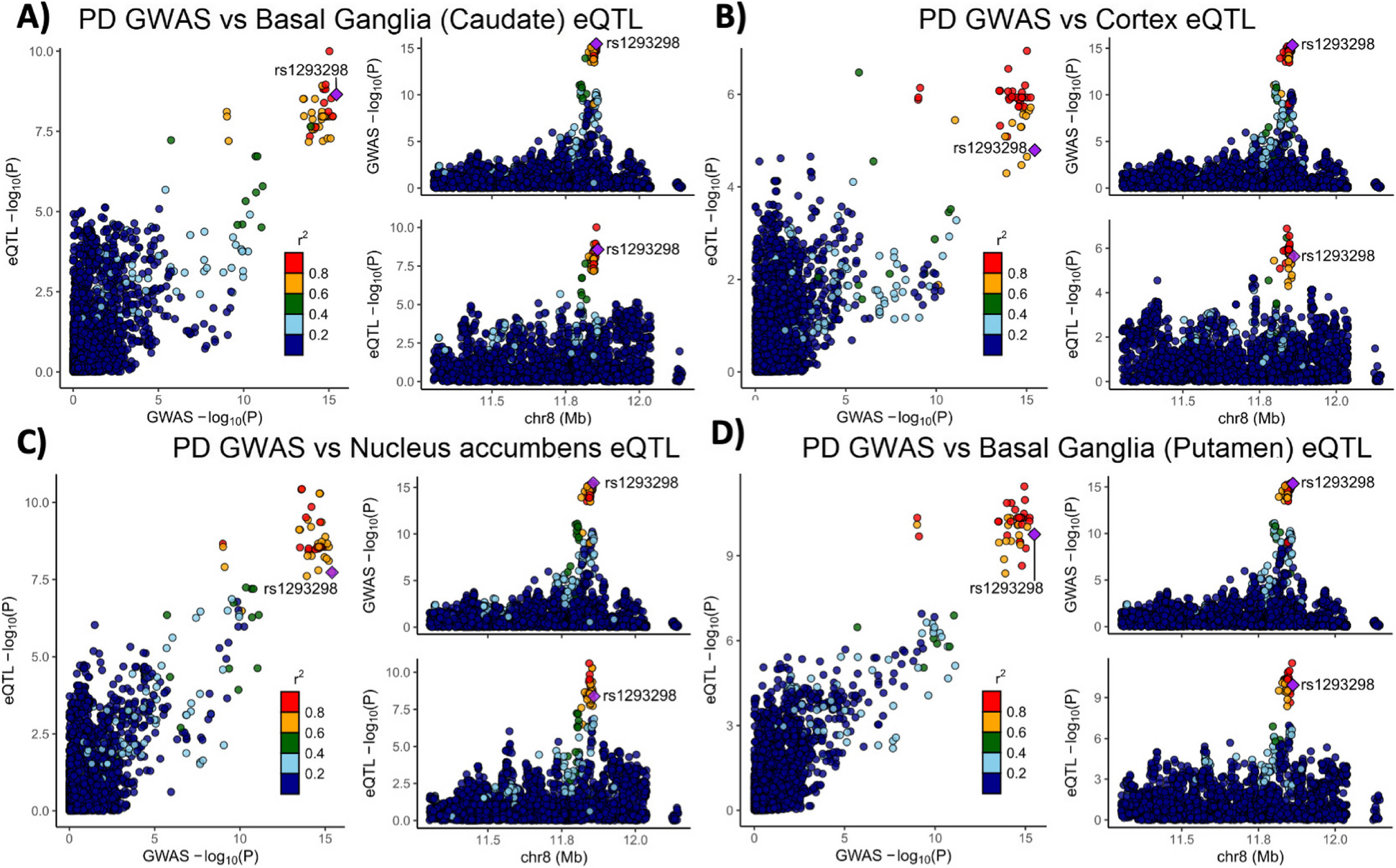
Variants in the genetic locus containing CTSB are significantly associated with risk of PD [9] yet this locus includes multiple other genes including FDFT1, NEIL2, GATA4 and it remains uncertain whether CTSB itself drives the association. We examined all the variants that are 1 MB upstream or downstream to the top GWAS variant in this locus and using GCTA-COJO, we show that an intronic CTSB variant (rs1293298, p = 3.41E-16, located in intron 1 of CTSB within a potential enhancer region) is the top variant associated with PD risk, without secondary associations. Fine mapping using FINEMAP gave this variant the highest posterior probability (0.127) of being causal, of all nominated variants. Since the top hit variant rs1293298 is located in a non-coding region, its impact is presumed to be on the regulation of gene expression rather than on the protein sequence. Indeed, this variant is in LD with multiple variants in this locus (r2 > 0.8) that are associated with CTSB expression with H4 posterior probability > 0.8 in multiple brain regions. The associations between genetic variants, PD and CTSB expression in PD-relevant brain regions such as basal ganglia, cortex and nucleus accumbens are depicted in Fig. 1. In particular, the minor allele of the rs1293298 CTSB variant linked to PD in GWAS exhibits a protective effect against PD and is associated with elevated expression levels of CTSB in brain tissues relevant to the disease (Fig. 1A-D). Analysis using SMR and HEIDI suggests that the QTLs in CTSB are potentially causally linked to PD with p HEIDI > 0.05 in multiple tissues (i.e., we could not reject the null hypothesis that there is a single causal variant affecting both gene expression and risk for PD). All results from the GCTA-COLOC, FINEMAP, SMR and HEIDI analyses are detailed in Supplementary Tables 9–11.
Genetic dissection of the CTSB locus in Parkinson’s disease risk. Locus zoom plots depicting the CTSB locus (± 500 kb) in Parkinson’s disease GWAS with brain eQTLs. The top PD-associated variant (rs1293298) is highlighted in purple, and variants in strong LD (r2 > 0.8) are in red. Each panel includes three plots: The left plot in each panel compares the p values from the PD GWAS and expression data for each variant. Variants that are in the top right corner of this plot are therefore associated with both risk of PD and CTSB expression. The top right plots depict the PD GWAS association in this locus and is identical in all four panels. On the bottom right of each panel, the plot depicts the association between variants in this locus and CTSB RNA expression in the relevant tissue. A PD GWAS plotted together with Basal ganglia (Caudate) eQTL. B PD GWAS plotted together with Cortex eQTL. C PD GWAS plotted together with Nucleus accumbens eQTL. D PD GWAS plotted together with Basal ganglia (Putamen) eQTL
These common CTSB variants occur in non-coding regions and likely exert their effects through altering expression levels. However, given the evidence that protective CTSB variants are associated with increased mRNA expression levels, we hypothesized that loss of function coding variants in CTSB would be likely to promote PD risk. We conducted rare variant analysis in 5,801 PD cases and 20,427 controls from six cohorts (Supplementary Table 2). We observed a nominal association between all rare variants and variants with high CADD score and PD risk in the Sheba cohort (p = 0.03 and p = 0.049, respectively). However, upon examining other cohorts and conducting a meta-analysis we did not find any additional associations (Supplemental Table 12). Additionally, we studied the role of rare CTSB variants on the age of PD onset. We found nominal association between functional variants and age at onset in the McGill cohort (p = 0.044) and in the meta-analysis for functional and non-synonymous variants (p = 0.048 and p = 0.043, respectively). All these results should be interpreted with caution as no p-values survived multiple comparisons.
CatB inhibition promotes α-syn aggregation in dopaminergic neurons
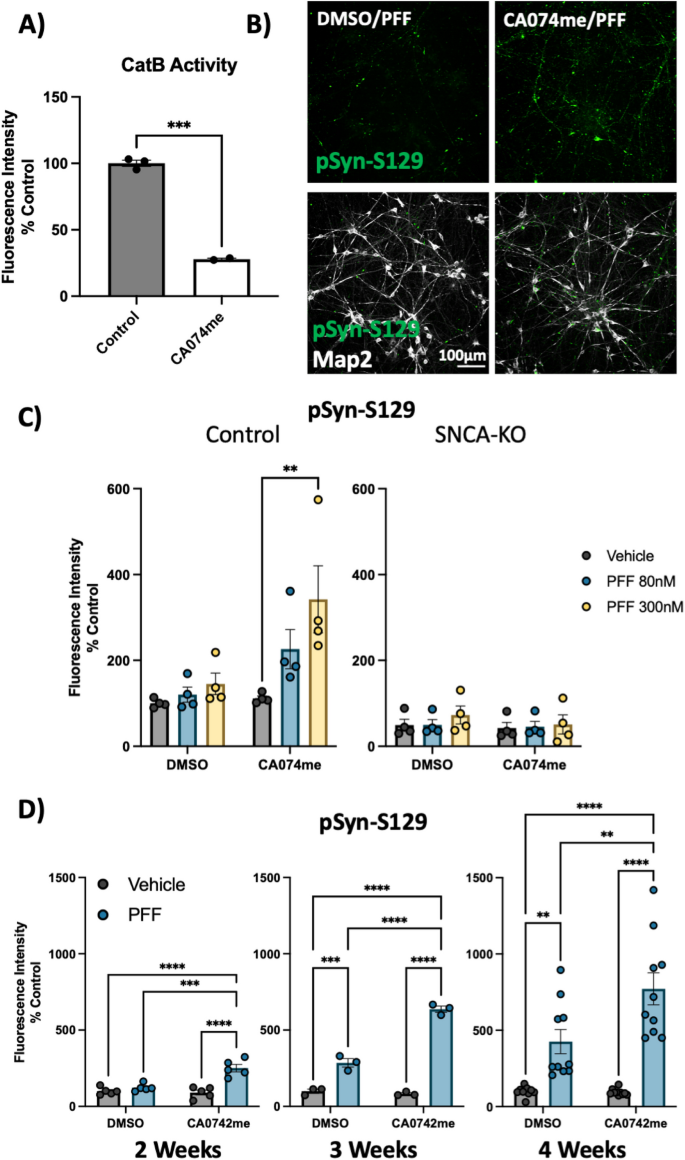
To functionally interrogate the role of catB in the handling of α-syn fibrils, we generated iPSC-derived DA neurons [41, 42] and treated them with pre-formed α-syn fibrils (PFFs) and the catB inhibitor CA074me [56] at a dose (1 μM), shown to significantly reduce catB activity (Fig. 2A). Exposure to PFFs promotes endogenous α-syn aggregation which can recapitulate many features of Lewy pathology including the accumulation of S129-phosphorylated α-syn (pSyn-S129) [57]. We used immunofluorescence and high-content confocal imaging to quantify pSyn-S129 using Map2 as a counterstain to define the region of neuronal cell bodies and proximal projections (Fig. 2B). At 3-weeks post-treatment PFF dose-dependently induced pSyn-S129 in DA neurons treated with CA074me but not vehicle (DMSO) (Fig. 2C). This pSyn induction was abolished in DA neurons lacking endogenous α-syn (SNCA-KO) (Fig. 2C), indicating that α-syn seeding, rather than phosphorylation of PFFs themselves gave rise to this pSyn-S129 signal. Next, using the 300 nM dose of PFF we tested a range of treatment durations and observed that after 2, 3 or 4 weeks, a single treatment with CA074me administered at the time of PFF exposure increased the abundance of pSyn-S129 (Fig. 2D). CA074me did not affect total α-syn levels (Fig S4A) and no loss of TH + DA neurons was observed 4 weeks post-treatment (Fig S4B). Similar potentiation of pSyn induction was observed with PADK, a distinct catB inhibitor (Fig S4C,D) reinforcing that this likely reflects on-target activity of catB inhibition.
Cathepsin B inhibitors potentiate the effect of α-syn PFFs on dopaminergic neurons: A) CatB activity measured by fluorogenic assay in DA neurons treated with 1 μM CA074me. B Representative immunofluorescent images from high-content confocal imaging of DA neurons treated with CA074me (1 μM) and/or α-syn PFFs (300 nM) and stained for Map2 and pSyn-S129. C Quantification of pSyn-S129 in Map2-positive cells 3-weeks after PFF and/or CA074me treatment in either Control (AIW002-2) DA neurons or isogenic neurons lacking endogenous α-syn (AIW002-2 SNCA-KO). D Quantification of pSyn-S129 in control DA neurons 2, 3, or 4 weeks after PFF (300 nM) and/or CA074me treatment. Bonferroni-corrected t-tests, ** p < 0.01, *** p < 0.001, **** p < 0.0001
CatB inhibition induces lysosome dysfunction in dopaminergic neurons
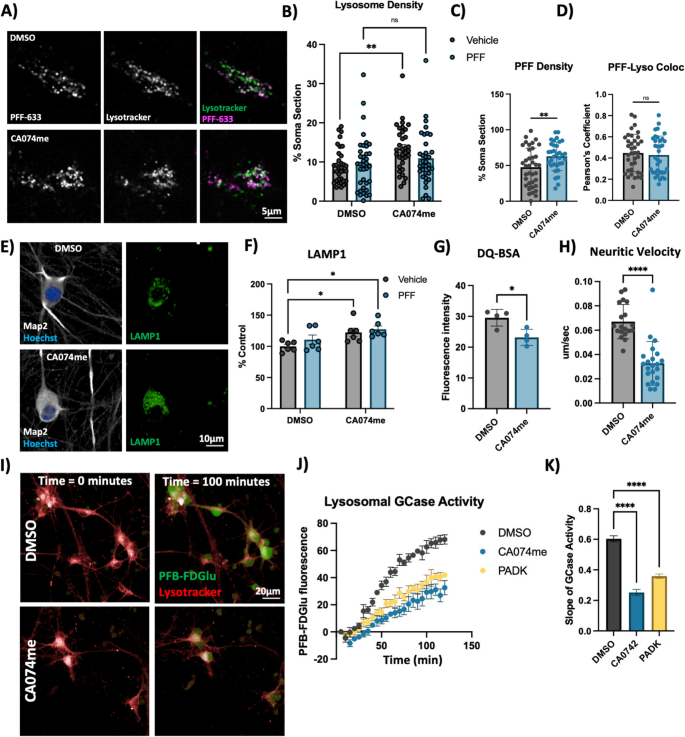
Extracellular α-syn aggregates are taken into neurons by a variety of mechanisms and are rapidly trafficked to lysosomes [58, 59]. To determine whether CA074me treatment altered the trafficking of PFFs into lysosomes or their persistence there, we performed live cell confocal imaging of DA neurons exposed to alexa-633 tagged PFFs (PFF-633) for 72 h and then stained with lysotracker (Fig. 3A). CA074me increased the overall lysosome content in cells not treated with PFF (Fig. 3B) and also increased the density of PFF-633 fluorescent puncta per cell (Fig. 3C) but colocalization of PFF-633 with lysosomes was unchanged (Fig. 3D). We interpret these observations as indicating that although the abundance of both lysosomes and PFF-633 within each cell is slightly elevated in CA074me treated neurons, the proportion of PFF-633 trafficked to lysosomes is unaffected.
Cathepsin B inhibition increases lysosome abundance but impairs function in dopaminergic neurons. A Representative live-cell confocal images of neuronal cell bodies stained with lysotracker-green 72-h after exposure to alexa-633 labelled α-syn PFFs (80 nM). B Lysosome density per cell body, measured as the percentage of lysotracker-positive area per cell soma. C PFF density per cell body, measured as the percentage of PFF-633-positive area per cell soma. D Colocalization of lysotracker and PFF-633 measured using Pearson’s coefficient per cell soma. E Representative immunofluorescent images from high-content confocal imaging of DA neurons treated with CA074me (1 μM) and/or α-syn PFFs (300 nM) for 3 weeks and stained for Map2 and LAMP1. F Quantification of LAMP1 in Map2-positive cells. G Lysosomal degradative capacity measured by fluorescence intensity of DQ-BSA fluorogenic probe 24-h after dye loading. H Quantification of lysosome velocity in neurites measured by live-cell confocal imaging and quantified using TrackMate. Points represent individual quantified image fields derived from 6 independent experiments. I Representative images of neurons stained with lysotracker deep-red and PFB-FDGlu fluorescent signal at baseline and 100 min after dye-loading. J Quantification of PFB-FDGlu fluorescence per cell in DA neurons pre-treated for 24-h with CA074me or PADK. K Quantification of the slope of PFB-FDGlu fluorescence versus time. T-test or Bonferroni-corrected t-tests, * p < 0.05, ** p < 0.01, **** p < 0.0001
Next, we further characterized how catB inhibition affected lysosome abundance and function. Similar to lysotracker, the abundance of the lysosomal membrane protein LAMP1 was increased after CA074me treatment, independent of concurrent PFF exposure (Fig. 3E, F). However, the degradative capacity of lysosomes (measured using the fluorogenic probe DQ-BSA) was reduced (Fig. 3G). The speed of lysosomal trafficking in neuritic projections was also reduced following CA074me (Fig. 3H). Lastly, given the genetic interaction between variants in CTSB and GBA1 in PD risk [10] and that catB has been found to regulate glucocerebrosidase (GCase) activity in HEK293 cells [60], we assessed the impact of catB inhibition on lysosomal GCase activity in DA neurons using the fluorogenic probe PFB-FDGlu (Fig. 3I-K). CatB inhibition with either CA074me or PADK impaired lysosomal GCase activity (Fig. 3J, K). These observations indicate that despite increasing lysosome abundance, catB inhibition impairs several aspects of lysosome function, including degradative capacity, trafficking and GCase activity.
To determine whether altered lysosome function could be related to the increased abundance of α-syn aggregates, (which has been found to impact lysosomal hydrolase trafficking [11, 13]), we differentiated 3xSNCA and SNCA-KO iPSCs [45] into DA neurons and treated them with CA074me for 3 weeks. We observed that while total levels of α-syn were unchanged by CA074me (Fig S5A,D), LAMP1 was increased in both 3xSNCA and SNCA-KO neurons (Fig S5B,E) indicating the increase in lysosome content is independent of α-syn. Additional staining using the Syn303 antibody, which has been reported to preferentially detect oxidized α-syn species which are often found in pathological aggregates [61], revealed that CA074me treatment increased Syn303 levels (Fig S5C,F) although no S129-pSyn signal above background was detected in these cells. Lastly we observed that CA074me impaired GCase activity in both 3xSNCA and SNCA-KO neurons (Fig S5G,H).
CTSB repression impairs autophagy and lysosomal function in RPE1 cells
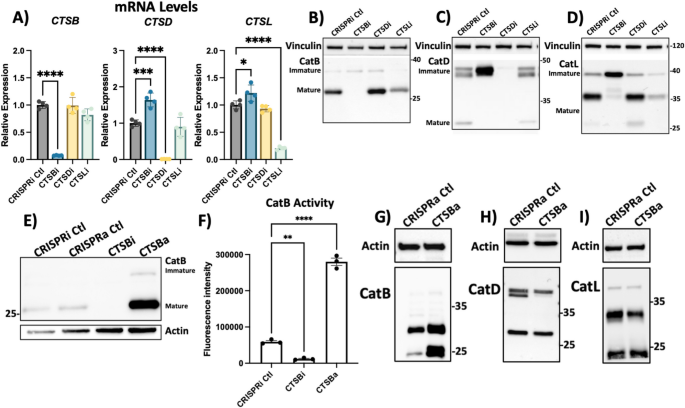
While CA074me is selective for catB, it has been reported to inhibit other cathepsins, albeit at concentrations greater than those used in this study [62, 63], including CTSD and CTSL which were previously found to cleave α-syn in vitro [18,19,20]. To determine how individual cathepsin species regulate lysosome function and contribute to fibrillar α-syn clearance we used CRISPR-interference (CRISPRi) to generate RPE1 cell lines in which CTSB, CTSD, CTSL or α-syn (SNCA) were stably repressed (denoted CTSBi, CTSDi, CTSLi and SYNi respectively) as well as CRISPR-activation (CRISPRa) to upregulate CTSB (CTSBa). Knockdown or upregulation of target genes was confirmed at transcript and protein levels (Fig. 4A-I). Unexpectedly, we observed that knockdown of CTSB resulted in increased mRNA levels (Fig. 4A), but impaired protein maturation of both catD and catL, indicated by increases in the higher molecular weight immature forms of the protein, and reduction in the proteolytically processed mature forms (Fig. 4C, D). Increased CTSB expression by CRISPRa was confirmed to lead to increased mature catB protein (Fig. 4E) and increased catB activity (Fig. 4F). CTSB activation also appeared to increase the protein levels of catD but not catL (Fig. 4G-I).
Generation of CTSB, CTSD and CTSL knockdown cell lines. A Validation of target knockdown in CRISPRi RPE1 cells. CTSB, CTSD and CTSL mRNA levels were measured by qPCR in control (CRISPRi Ctl), CTSB-knockdown (CTSBi), CTSD-knockdown (CTSDi) and CTSL-knockdown (CTSLi) cell lines. B CatB protein levels in control, CTSBi, CTSDi and CTSLi cell lines. C CatD protein levels in control, CTSBi, CTSDi and CTSLi cell lines. D CatL protein levels in control, CTSBi, CTSDi and CTSLi cell lines. E Protein levels of CatB in CRISPRi control, CRISPRa control, CTSB knockdown (CTSBi) and CTSB upregulation (CTSBa) RPE1 cell lines. F CatB activity measured by fluorogenic assay. G–I CatB, CatD and CatL protein levels in control and CTSBa cells
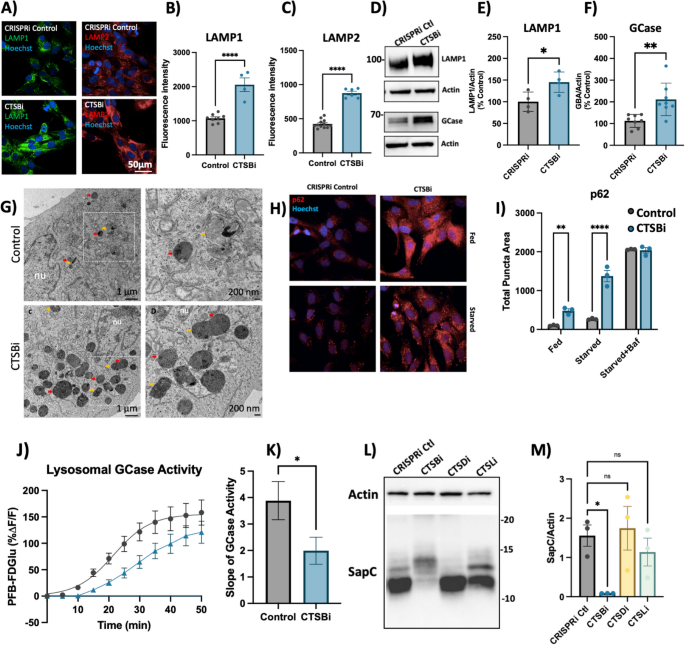
Similar to what we observed in CA074me treated DA neurons, we found that CTSBi cells had significantly increased abundance of LAMP1 and LAMP2 immunofluorescent puncta (Fig. 5A-C), increased LAMP1 and GCase protein levels (Fig. 5D-F) and increased number and size of electron dense lysosome-like structures (including lysosomes and multivesicular bodies) observed by electron microscopy (Fig. 5G). To determine whether impairment in autophagic flux could contribute to the accumulation of lysosomes, we measured the abundance of p62 puncta under fed and starved conditions, and in the presence of bafilomycin (to inhibit lysosomal clearance of autophagosomes). CTSBi resulted in increased abundance of p62 puncta under fed and serum-starved conditions, but not in the presence of bafilomycin (Fig. 5H, I) suggesting an impairment in the clearance of autophagosomes. Similarly, we observed accumulation of the autophagy-associated proteins LC3B and p62 by western blot in CTSBi cells (but not CTSDi or CTSLi), in the absence of serum starvation (Fig S6A,B).
CTSB repression increases lysosome abundance but impairs function in RPE1 cells. A–C Representative images and quantifications of LAMP1 and LAMP2 immunofluorescence in CRISPRi control and CTSBi RPE1 cells. D–F Representative western blots and quantifications depicting protein levels of LAMP1 and GCase/GBA relative to actin. G Electron microscopy images of CRISPRi and CTSBi RPE1 cells. Electron-dense multivesicular bodies-like structures are indicated by red asterisk, lysosomes by orange asterisk, secondary lysosomes by green asterisk, whereas mitochondria are indicated by blue asterisk. H, I Representative immunofluorescent images and quantification of p62 area per cell in RPE1 cell lines (CRISPRi control – grey bars, and CTSBi – blue bars) under fed, 16-h starvation and 16-h starvation with bafilomycin conditions. J GCase activity measured as PFB-FDGlu fluorescence intensity over time. K Quantification of the slope of the PFB-FDGlu fluorescence versus time curves. L, M Representative saposin C (sapC) western blot and quantification relative to actin loading control. Bonferroni-corrected t-tests, * p < 0.05, *** p < 0.001. T-test or Bonferroni-corrected t-tests, * p < 0.05, ** p < 0.01, **** p < 0.0001
Despite this increase in lysosome content and increased GCase protein levels (Fig. 5D, F), the activity of lysosomal GCase per cell was significantly reduced by CTSB knockdown (Fig. 5J, K). One potential mechanism linking CatB to GCase activity is via the cleavage of pro-saposin into saposin C (SapC), which acts as a co-activator of GCase [60]. Using an antibody that recognizes the SapC fragment of the pro-saposin peptide (PSAP) we found that in CTSBi, but not CTSDi or CTSLi cells, the levels of SapC were significantly reduced (Fig. 5L, M), while uncleaved pro-saposin species were increased (Fig S6C). This supports a prior finding that CatB is required for PSAP processing into SapC [60] and may contribute to the reduction in GCase activity.
In addition to impaired clearance of autophagosomes, a second factor that could contribute to increased lysosome abundance is increased lysosome biogenesis. In fact catB has previously been reported to regulate activation of the transcription factor TFEB [64, 65]. Given that mRNA levels of CTSD and CTSL were already noted to be increased in CTSBi cells (Fig. 4A), we suspected a potential role for increased TFEB activity. Indeed, we found that in CTSBi cells, nuclear localization of TFEB was increased in non-starved cells (Fig S6D, E) and mRNA levels of several lysosomal genes (including LAMP1, GBA and MCOLN1) were transcriptionally upregulated (Fig S6F). These results taken together indicate that a combination of impaired lysosomal enzyme activity, impaired lysosome turnover and upregulated lysosome biogenesis contribute to the increased abundance of dysfunctional lysosomes in RPE1 cells lacking CTSB.
CTSB levels regulate PFF clearance in RPE1 cells
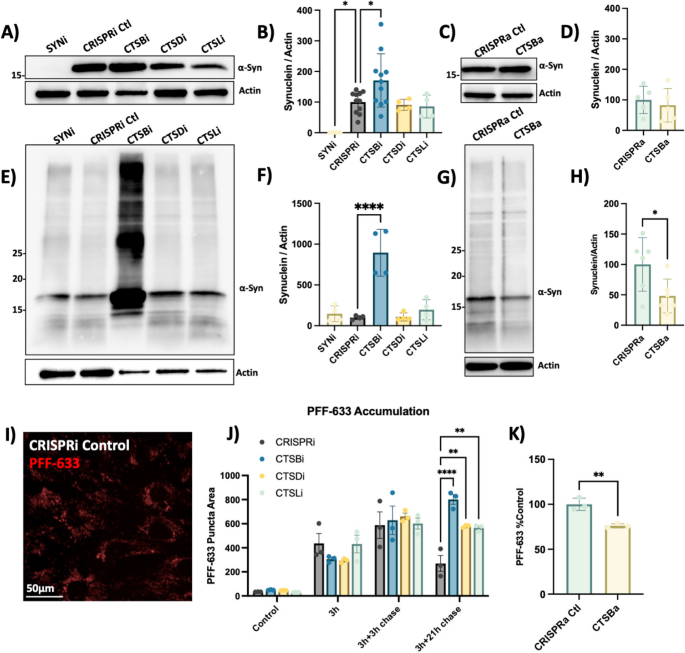
We next examined the effect of cathepsin knockdown on α-syn protein levels. Endogenous α-syn protein was undetectable in SNCAi cells and elevated in CTSBi cells (Fig. 6A, B). In contrast, CTSBa had no effect on endogenous α-syn protein (Fig. 6C, D). Strikingly, when these cells were exposed to exogenous PFFs for 48 h, CTSBi cells (but not CTSDi or CTSLi) exhibited significantly increased abundance of α-syn, including high molecular weight species, compared to control cells (Fig. 6E, F). Conversely, CTSBa had the opposite effect, modestly reducing the levels of aggregated α-syn (Fig. 6G, H). Moreover, increased α-syn levels were also observed in control or SNCAi cells treated with CA074me (Fig S7) indicating that this increase reflects either increased cellular uptake or failed clearance of the PFFs, rather than new aggregate seeding.
CTSB but not CTSD or CTSL repression impairs PFF clearance in RPE1 cells. A Representative western blot depicting protein levels of α-syn and actin. B Western blot quantification depicting protein levels of α-syn relative to actin expressed as percentage of CRISPRi control. C Representative western blot depicting protein levels of α-syn and actin. D Western blot quantification depicting protein levels of α-syn relative to actin expressed as percentage of CRISPRa control. E Representative western blot depicting protein levels of α-syn and actin 48-h after treatment of RPE1 cell lines with 300 nM of α-syn PFFs. F Western blot quantifications depicting levels of α-syn (quantification of whole lane) relative to actin in PFF treated RPE1 cells. G Representative western blot depicting protein levels of α-syn and actin 48-h after treatment of RPE1 cell lines with 300 nM of α-syn PFFs. H Western blot quantifications depicting levels of α-syn (quantification of whole lane) relative to actin in PFF treated RPE1 cells. I Representative image of CRISPRi control RPE1 cells 48-h after treatment with alexa-633 tagged α-syn PFFs (80 nM). J, K Quantification of PFF-633 fluorescent intensity per cell in RPE1 cell lines. T-test or Bonferroni-corrected t-tests, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001
To determine whether the uptake or clearance of PFFs was affected, we used Alexa-633 fluorescently labelled PFFs and conducted a pulse-chase experiment to monitor the uptake and subsequent clearance of PFFs (Fig. 6I, J). During a 3 h exposure, or 3-h exposure with short washout (3 h chase), the PFF-633 levels per cell were similar across cell lines. However, when washout was extended to 21-h CTSBi cells retained more PFF-633 (Fig. 6J), suggesting impaired clearance. CTSDi and CTSLi also appeared to impair PFF clearance in this assay but to a lesser extent than CTSBi. Similar to what we observed by Western blot, when we exposed CTSBa cells to PFF-633 for 48 h, we observed a reduced accumulation of the tagged PFF (Fig. 6K). Taken together, these findings indicate that CatB regulates the clearance of internalized α-syn aggregates, though this may represent either a direct role for CatB in α-syn degradation, or an indirect effect secondary to broader lysosome impairment under conditions of CTSB depletion.
Knockout of CTSB in human dopaminergic neurons leads to lysosomal dysfunction
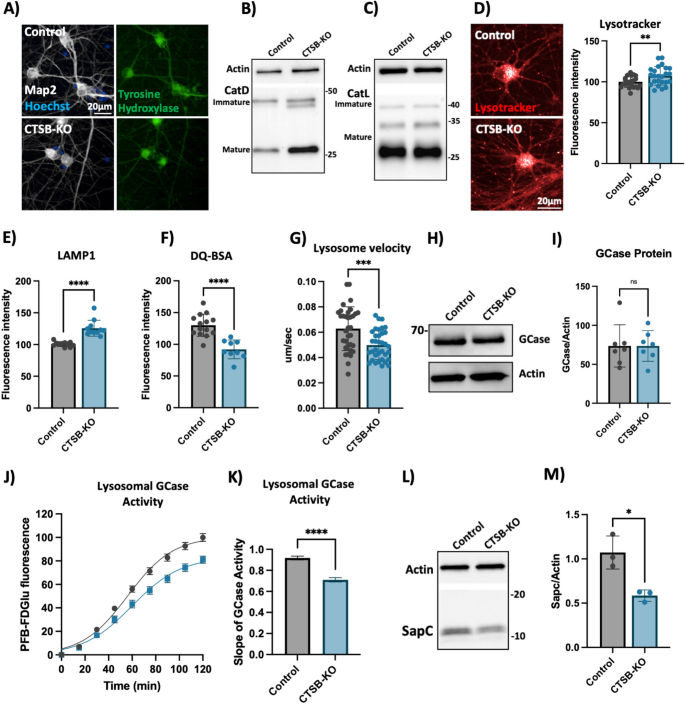
To further confirm the lysosomal phenotypes that we previously observed in neurons treated with CA074me, we generated CTSB knockout iPSCs (CTSB-KO) and differentiated them into DA neurons (Fig. 7A). Both control and CTSB-KO neurons exhibited similar expression patterns of dopaminergic genes (Fig S8A-D), and produced similar percentages of TH-positive neurons within each differentiation (Fig S8E), indicating that loss of CTSB did not impact the ability of iPSCs to differentiate into DA neurons. In contrast to CTSBi RPE1 cells, CTSB-KO DA neurons did not exhibit an impairment of CatD or CatL protein processing (Fig. 7B, C). However, similar to CA074me treatment, in CTSB-KO neurons lysosome abundance measured either by lysotracker (Fig. 7D) or LAMP1 immunofluorescence (Fig. 7E) were increased, while degradative capacity (Fig. 7F) and neuritic trafficking velocity (Fig. 7G) were reduced. GCase protein levels were unaffected (Fig. 7H, I) however lysosomal GCase activity was reduced by 20% in CTSB-KO neurons (Fig. 7J, K). We also found that similar to CTSBi RPE1 cells, levels of SapC were significantly reduced in CTSB-KO neurons, which could contribute to the reduction in GCase activity (Fig. 7L, M). Lastly, unlike CTSB-knockdown RPE1 cells, CTSB-KO neurons did not exhibit detectable activation of TFEB (Fig S8F) or transcriptional upregulation of lysosomal genes (Fig S8G) arguing that CatB may regulate TFEB activity and lysosome biogenesis in a cell-type or context-dependent manner.
CTSB knockout impairs lysosome function in dopaminergic neurons. A Representative immunofluorescent images from high-content confocal imaging of iPSC-derived DA neurons differentiated from control or CTSB-KO iPSCs and stained for Map2, tyrosine hydroxylase (TH) and α-syn. B Representative western blot depicting CatD protein levels in control and CTSB-KO neurons. C Representative western blot depicting CatL protein levels in control and CTSB-KO neurons. D High-content imaging representative image and quantification of lysotracker fluorescence in DA neurons. E LAMP1 immunofluorescence per cell in Map2-positive DA neurons. F Lysosomal degradative capacity measured by fluorescence intensity of DQ-BSA fluorogenic probe 24-h after dye loading. G Quantification of lysosome velocity in neurites measured by live-cell confocal imaging and quantified using TrackMate. Points represent individual quantified image fields derived from 6 independent experiments. H, I Representative western blot and quantification of GCase and actin in DA neurons. J Quantification of PFB-FDGlu fluorescence per cell in DA neurons K) Quantification of the slope of PFB-FDGlu fluorescence versus time. L, M Representative saposin C (sapC) western blot and quantification relative to actin loading control. T-tests, ** p < 0.01, *** p < 0.001, **** p < 0.0001
CTSB deficiency promotes synuclein pathology in human dopaminergic neurons and midbrain organoids
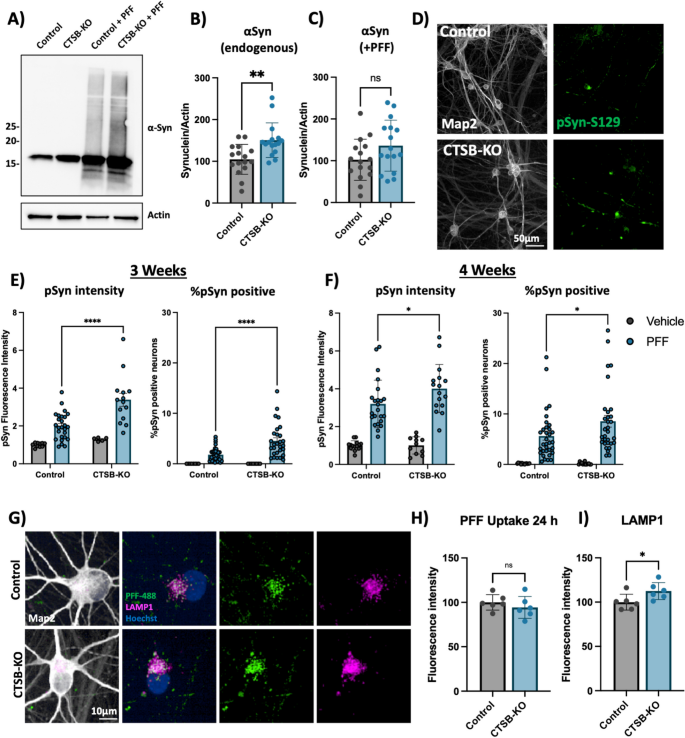
Compared to parental control, CTSB-KO DA neurons were found to have modestly elevated levels of endogenous α-syn (Fig. 8A,B). When treated with PFFs for 72 h CTSB-KO neurons did not exhibit higher levels of total α-syn (Fig. 8A, C). However, 3 and 4 weeks after PFF treatment CTSB-KO neurons accumulated significantly more pSyn-S129, and this was evident when measuring either the average pSyn-S129 intensity within Map2-positive neurons, or the percentage of pSyn-positive cell bodies (Fig. 8D-F). The efficiency of PFF uptake (measured by alexa-488 tagged PFF internalization) was unaffected in CTSB-KO neurons (Fig. 8G,H) but as expected LAMP1 was elevated (Fig. 8G,I). Using live-cell confocal microscopy, we observed a modest increase in total PFF-633 levels in CTSB-KO neurons 72 h after treatment (Fig S9A, B), however there was no difference in the trafficking of alexa-633 tagged PFFs to lysosomes as measured by PFF-633 and lysotracker colocalization (Fig S9C). To explore whether CTSB overexpression, or re-expression in the CTSB-KO background could reduce PFF-induced pSyn-S129, we transduced DA neurons with an adeno-associated viral vector (AAV) expressing CTSB along with the fluorescent protein amCyan (Fig S9D). We observed a significant reduction in pSyn-S129 puncta in the CTSB-KO neurons transduced with AAV-CTSB compared to AAV-GFP control (Fig S9E), despite modest levels of CTSB expression achieved with the AAV transgene (Fig S9F), arguing that even small amounts of CTSB expression can promote pSyn-S129 clearance.
CTSB knockout enhances the effect of α-syn PFFs on dopaminergic neurons. A Representative western blot showing levels of α-syn in untreated control or CTSB-KO DA neurons as well as PFF treated DA neurons. B, C Western blot quantifications depicting protein levels of α-syn relative to actin for endogenous α-syn (B) or PFF (C), normalized to the respective control. D Representative immunofluorescent images from high-content confocal imaging of Control or CTSB-KO DA neurons treated with α-syn PFFs (300 nM) and stained for Map2 and pSyn-S129. E, F) Quantification of pSyn-S129 in Map2-positive cells 3-weeks (E) and 4-weeks (F) after PFF treatment. Left graphs depict pSyn-S129 fluorescence intensity within Map2-positive cells, and right graphs depict the percentage of Map2-positive cell bodies positive for pSyn-S129 aggregates. G Representative immunofluorescent images from high-content confocal imaging of DA neurons treated with alexa-488 tagged PFFs (PFF-488, 80 nM) for 24 h and stained for LAMP1 and Map2. H Quantification of PFF-488 fluorescence per Map2-positive cell. I Quantification of LAMP1 fluorescence per Map2-positive cell. T-tests or Bonferroni-corrected t-tests, ** p < 0.01, **** p < 0.0001
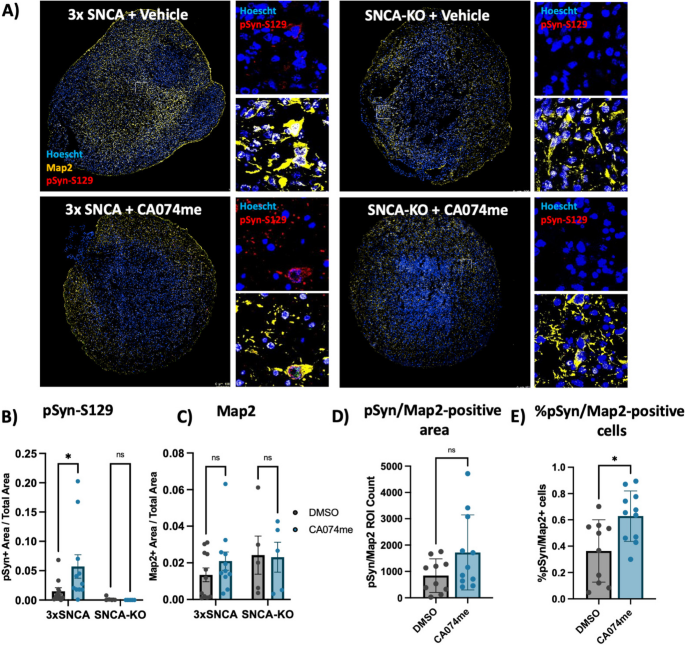
To determine whether the loss of catB function could promote α-syn aggregation independent of PFF exposure, we generated midbrain organoids from patient-derived iPSCs harboring an SNCA triplication mutation (3xSNCA) and an isogenic SNCA-KO line (Fig S3). We have previously described the characterization of these 3xSNCA organoids that spontaneously develop pSyn-S129-positive α-syn aggregates after sustained culture [45]. To determine whether endogenous α-syn aggregation would be affected by catB impairment we treated 3xSNCA or SNCA-KO organoids for 60 days with DMSO (vehicle) or 1 μM CA074me. We observed an increase in the abundance of pSyn-S129 (measured as the area positive for pSyn-S129 relative to total organoid area) (Fig. 9A,B), while treatment had no effect on total neuron content (Map2-positive area) (Fig. 9C). We also observed a trend towards an increase in the total Map2/ pSyn-S129 colocalized area (Fig. 9D), and an increase in the percentage of Map2-positive cells that were positive for pSyn-S129 (Fig. 9E). Co-staining of pSyn-S129 with TH revealed that pSyn signal originated partially, but not exclusively from dopaminergic neurons in both vehicle and CA074me treated organoids (Fig S10). This is similar to what is observed in postmortem PD brain, where pathological inclusions are not limited to dopaminergic neurons [66], and further indicate that loss of catB function increases the burden of pathological α-syn aggregates.
CTSB inhibition promotes pSyn-S129 accumulation in patient-derived midbrain organoids. A Representative immunofluorescent images of Map2 and pSyn-S129 in 5-month old SNCA-triplication (3xSNCA) and isogenic SNCA-KO midbrain organoids treated with vehicle (DMSO) or 1 μM CA074me for 60 days. Large images depict representative whole-organoids and high-magnification images depict individual Map2-positive cells. B Quantification of the pSyn-S129 positive area of the organoid relative to the total organoid size. C Quantification of the Map2-positive area relative to organoid size. D Quantification of pSyn-S129- and Map2 double-positive area in 3 × SNCA organoids. E Quantification of the percentage of cells positive for both Map2 and pSyn-S129 in 3 × SNCA organoids. Bonferroni-corrected t-tests, * p < 0.05